CERTISAI PREDICTIVE ONCOLOGY INTELLIGENCE™
CertisAI™ is the first and only patented, commercially available, validated AI/ML platform that predicts therapeutic efficacy by identifying complex gene expression signatures that correlate with drug response. The platform accelerates drug discovery and development by bringing computer-aided precision to IND-enabling, drug repurposing, and label expansion initiatives:
-
- Select optimal cancer models for in vitro and in vivo validation studies
- Identify gene expression signatures that are predictive of therapeutic response or resistance
- Derive insights into drug mechanisms of action
- Rank cancer types most likely to respond to a lead candidate
- Elucidate synergistic potential of combinatory therapies
- Develop stratification strategies for clinical trial patient selection
- Prioritize candidates in compound libraries
Leveraging the computational power of CertisAI, cancer researchers can effectively replace many discovery screening studies with insights derived from in silico models.
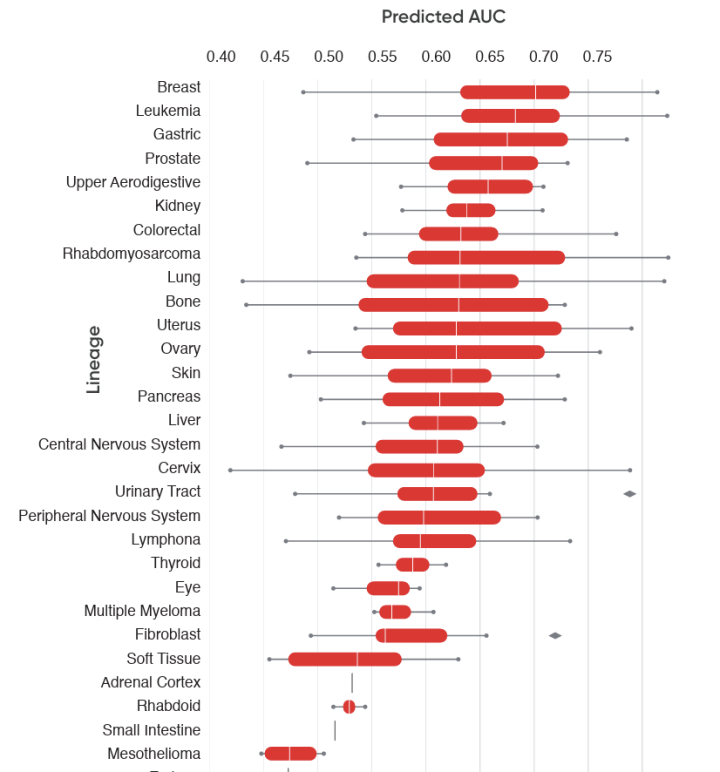
Informing Commercial Strategy
Here, CertisAI provides insights to inform a label expansion strategy by predicting PDX models’ response to an FDA-approved drug across cancer types.
WHY GENE EXPRESSION?
CertisAI predictions are based on drug features and the related gene expression profiles of responsive and non-responsive cancer models. Using gene expression as a predictive biomarker is far more nuanced than genetic mutations alone. It leverages the full dynamic range of close to 20,000 genes—all with continuous values of expression. This approach also captures functional gene activity, epigenetic and dynamic genomic changes, and pathway disruptions, offering a systems biology perspective that can be more informative for predicting how a tumor will respond to a particular therapy.
When generating molecule-specific custom CertisAI models, the platform typically identifies five to 20 genes of interest and their relative expression levels—a gene expression signature that is predictive of drug response and/or drug resistance.
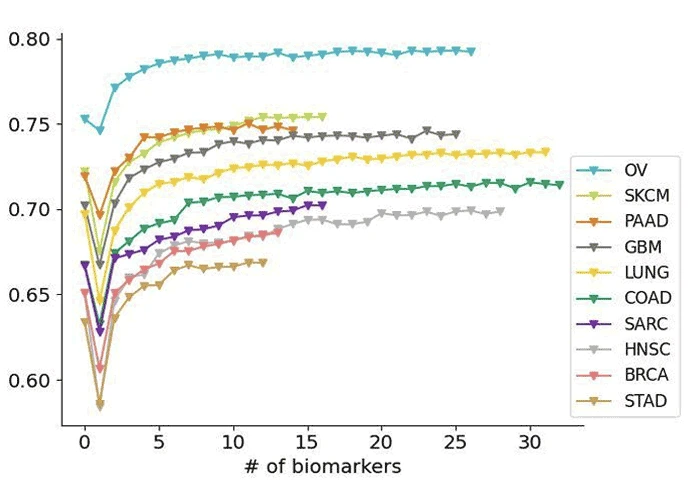
Expedite Drug Discovery and Development
In studies aimed at evaluating CertisAI performance, we discovered diminishing returns as drug features were combined with increasing numbers of selected gene biomarkers. Here, performance of monotherapy models is evaluated with five-fold cross validation (z-score R2).
MULTIVARIATE ANALYSIS: FROM BIG DATA INTO ACTIONABLE INSIGHTS
The development of CertisAI involved the integration of several proprietary and publicly available genomic, transcriptomic and drug response datasets. These data were integrated into a unified training framework through careful mapping, preprocessing, and transformation steps to ensure consistent, high-quality inputs.
Various machine learning algorithms including random forest models, support vector machines, and neural networks, were used to train CertisAI to recognize complex relationships between drug features and the gene expression signatures that correlate with drug response. This pre-built, multi-dimensional, relational map enables CertisAI to predict drug response not only for FDA-approved drugs in the training set, but also for investigational therapies the platform has never before encountered.
AI-Powered Insights for Precision Oncology
CertisAI encompasses a pre-built library of 3.5 million monotherapy predictions for 8,000 FDA-approved and investigational drugs, and 1.9 billion combination therapy predictions for 2,200 FDA-approved drugs. All of Certis’ proprietary PDX and PDX-derived models, as well as the nearly 1,400 models in the Cancer Cell Line Encyclopedia (CCLE) are represented.
PRETRAINED VS CUSTOM MODELING
CertisAI can be leveraged across a broad range of cancer therapeutics, from small molecules to peptides, monoclonal antibodies (mAbs), bispecific antibodies (BsAbs), and antibody-drug conjugates (ADCs).
Predictions from pre-trained CertisAI models require only a small molecule drug structure or Simplified Molecular Input Line Entry System (SMILES) string. For other therapy types, we leverage the CertisAI platform to build molecule-specific, custom AI models using response data from previously conducted in vitro or in vivo screening studies. Area Under the Curve (AUC) data are preferred, but other relevant endpoints such as cell viability, IC50 or EC50, or TGI data may be used.
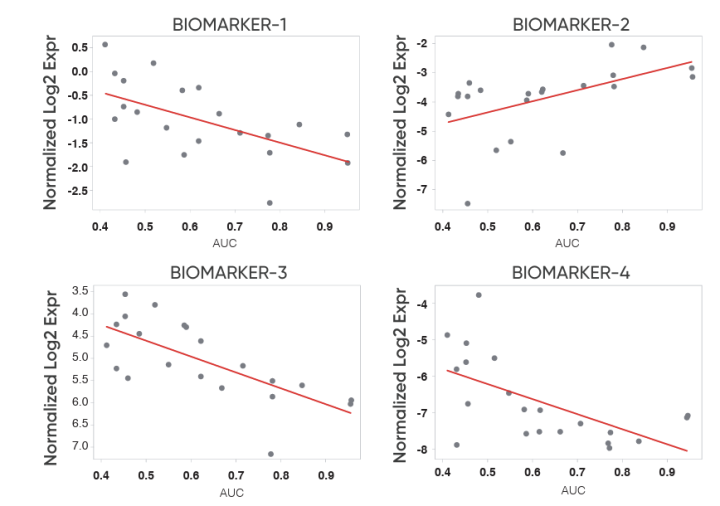
Efficacy Predictions on Certis PDX Models
Efficacy prediction on Certis PDX models using predictive biomarkers from a drug’s custom AI model. Custom CertisAI models are specific to individual therapies and are typically refined over time with additional training data from in vitro and/or validation studies.
YES, CERTISAI IS VALIDATED
Unlike many artificial intelligence/machine learning platforms, CertisAI has been validated—both internally using in silico methods, and externally via in vitro and in vivo pharmacology experiments.
Internal validation, also known as cross-validation, is a machine learning validation method that assesses an AI model’s performance. Certis data scientists employed a “hold-out” approach, whereby the platform is trained using 80% of the full dataset; the remaining 20% is used to test the accuracy of predictions. When predictions were correlated with actual AUC seen in the hold-out data, CertisAI predicted monotherapy response with 90% accuracy, and combination drug response with 80% accuracy.
External validation of the platform is an ongoing process. In vitro and in vivo pharmacology studies performed to date consistently demonstrate a strong positive correlation between actual and predicted tumor growth inhibition (TGI).
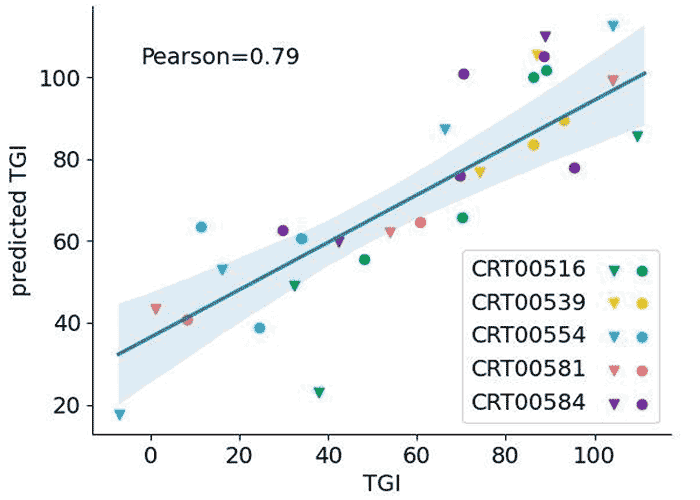
Validating Predictions: TGI Accuracy in Colorectal Studies
Shown here is predicted vs. actual TGI from five O-PDX colorectal pharmacology studies. Each data point represents a drug response; monotherapy data points are denoted by inverted triangles, drug combinatorial therapies shown as circles. Accuracy is evaluated using the Pearson coefficient, in which correlations above .7 are generally considered to be strong.
CertisAI: Answers to Frequently Asked Questions
How does Certis ensure the security of proprietary client data?
How does CertisAI predict the efficacy of oncology drug synergies?
How can CertisAI support label expansion or drug repurposing efforts?
How much do CertisAI services cost?
What are the clinical applications of CertisAI?
Learn more about leveraging artificial intelligence in cancer research
Download the CertisAI factsheet